Welcome to GlycoEnzDB
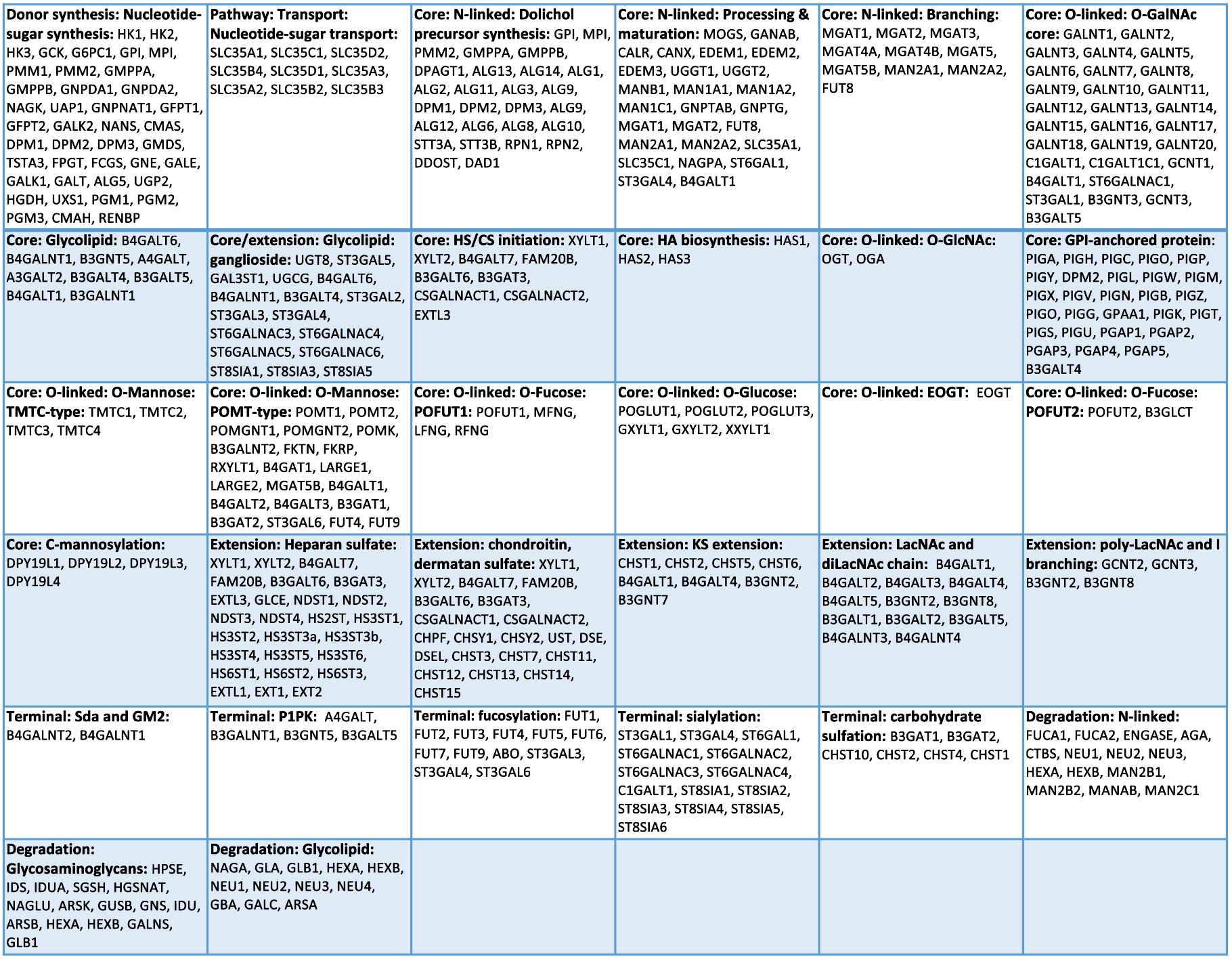
Gene Pathway Map (Clickable Blocks)
Licensing: CC BY 4.0. You are fee to copy, redistribute, remix, transform and build upon all material, except for textbook figures from the Essentials.