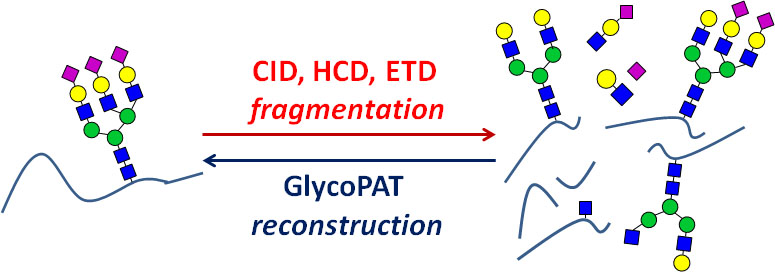
GlycoPAT is a modular, open-source MATLAB based toolbox that can be used for shotgun glycoproteomics data analysis (Version 1.0, released on September 8, 2017). This program provides a novel platform for the streamlined analysis of traditional LC-MSn based high-throughput experimental data for the identification of site-specific N- and O-linked glycosylation on various proteins.
Citation
Gang Liu, Kai Cheng, Chi Y. Lo, Jun Li, Jun Qu and Sriram Neelamegham. “ A comprehensive, open-source platform for mass spectrometry based glycoproteomics data analysis”, Molecular and Cellular Proteomics, 2017 Nov;16(11):2032-2047. Link
Since the original release, several critical improvements were made to the original GlycoPAT software as part of the HUPO Glycoproteomics first study, discussed elsewhere. Unfortunately, due to challenges associated with the pandemic these updates could not be released.
However, these updates have been incorporated into pyGlycoPAT, a python based glycoproteomics tool, built partly on the OpenMS and pyOpenMS frameworks. We hope to release this work shortly (manuscript in preparation, 2025).
Description
The unique features of the suite of programs include:
- The “SmallGlyPep” (SGP1.0) nomenclature: This nomenclature defines a minimal representation of glycopeptides in linear text format. It enables facile in silico glycan and peptide backbone fragmentation during MSn data analysis.
- Ensemble score (ES-score): This single parameter scores the glycopeptide hits based on cross-correlation analysis, Poisson probability and decoy database based calculated P-values, and additional considerations. Additionally, an acceptable ES-score for any experiment is calculated based on false discovery rate (FDR) calculations.
- User-extensible modular environment: The program provides a comprehensive set of functions for mass spectrometry data handling, computing glycopeptide mass/formula/fragmentation, glycopeptide digestion, spectra scoring, and MS/MS spectrum annotation. These functions are modular and thus they can be used in additional programs independent of GlycoPAT.
Availability
Freeware can be downloaded from glycopat.sourceforge.net
Operating system: Program is platform independent and designed to work on Windows, Mac and Linux machines.
Source code: Fully documented source code is provided at the above repository.
The flavors of GlycoPAT: The program can be run using three methods:
- Standalone application: Here, the user downloads the GUI (Graphical User Interface) version that is already complied in different OS from sourceforge, and double-clicks to install *. Please follow the instructional videos below for detailed steps on how to use the program after installation. This version will be helpful to typical experimentalists since it does not require knowledge of programming or any additional software.
- Standalone application run from the MATLAB command line: This is identical to the above standalone application, only it is started at the MATLAB command prompt. This version will be useful for individuals who have access to the MATLAB software (version 2013b or later), and those who are thinking about adding specific functions to the GlycoPAT core code.
- Command line version: This is the most versatile form of GlycoPAT, particularly for those who want to send GlycoPAT jobs to computing clusters. Here, users can write scripts to automate a number of core functions already available in the program suite.
Complete Program Documentation: Full description and usage instructions are provided in the GlycoPAT_Manual.pdf file.
Instructional Videos: These videos describe the SGP1.0 (Small GlycoPeptide 1.0) nomenclature ; and the usage of the GUI (Graphical User Interface) version of the software.
* Note: To install PC and Mac versions of GlycoPAT, simply download and double-click to install. For the Linux version go to the installation folder; type: sudo ./InstallGlycoPAT_Linux.install and follow on-screen instructions. The default installation directory is: /usr/GlycoPAT/. The default location of the MCR (MATLAB Compiler Runtime) library is /usr/local/MATLAB/MATLAB_Compiler_Runtime/v90/.Following installation, add the following values to the environment variable
LD_LIBRARY_PATH:
{MCR_ROOT}/runtime/glnxa64
{MCR_ROOT}/bin/glnxa64
{MCR_ROOT}/sys/os/glnxa64
{MCR_ROOT}/sys/opengl/lib/glnxa64
Here ‘{MCR_ROOT}’ is the MCR library path. After the environment variable is set, go to the GlycoPAT installation folder. Navigate into its subfolder “application”. Run the shell script as ./run_GlycoPAT.sh {MCR_Root}. This will launch the program.
Standalone version
Some modules of the GlycoPAT program have been complied with a graphical user interface (GUI) for easy use by any experimentalist. This version can be launched by simply downloading the software from glycopat.sourceforge.net and double-clicking to install (see *Note above). No other ancillary software is necessary to launch this standalone application.
As an option, the standalone application can also be triggered from the MATLAB command prompt.
This version of the program will run on a local computer, and it follows a three-step workflow that includes:
- The in silico generation of a library of putative glycopeptides by protease digestion.
- Tandem MS analysis using a novel MS scoring scheme to make glycopeptide identifications.
- A spectrum and fragmentation viewer to visualize and annotate the results.
Additional GUIs are also available for the manual refinement of single MS spectrum annotations, and for performing theoretical in silico glycopeptide fragmentation.
Command line version
Full program source files are provided so that all the features of the GlycoPAT program can be implemented using MATLAB command line operations or by writing scripts. Extensive help documentation including examples are provided with the code for users more familiar with programming who wish to integrate selected or all features of the GlycoPAT program into their own software. This version of MATLAB is also ideal for individuals wishing to run GlycoPAT on super-computing clusters. The functions in this version are broken down into those that handle:
- GUIs
- Mass spectrometry data handling
- Glycopeptide digestion
- Glycopeptide fragmentation
- Glycopeptide mass calculations
- Generation of decoys and false discovery rate (FDR) calculations
- Spectrum scoring
- High performance computing
Frequently asked questions
What platform will the program run on?
The program operates on PC, Mac and Linux systems. It is platform independent.
Is this program being further developed?
Yes, a python version of the code is being developed as of summer, 2025. This is being tested and will be released as soon as possible.
I need more help?
Look at the sourceforge site that contains code and test examples (http://glycopat.sourceforge.net), and also the github site for raw code (https://github.com/kaichengub/GlycoPAT). The contact for this project is neel@buffalo.edu (Sriram Neelamegham).