A program for the detailed annotation of MS/MS glycoproteomics spectra using DrawGlycan-SNFG sketches
Citation: Cheng et al. ‘DrawGlycan-SNFG & gpAnnotate: Rendering glycans and annotating glycopeptide spectra’, Bioinformatics, 36(6):1942-43, 2020 [link].
Links to: View User Manual
Download user manual and test data set [.zip file].
Download Compiled Program for Win/Mac/Linux. To install, download and double-click (disable anti-virus software, if necessary)
About gpAnnotate
The Glycopeptide Spectrum Annotation program (gpAnnotate) is designed to annotate individual glycoproteomics MS/MS spectrum for several established mass spectrometry fragmentation modes:
- CID: collision induced dissociation
- HCD: beam-type CID or higher-energy collision dissociation
- ETD: electron transfer dissociation
- ETciD: electron-transfer-CID, and
- EThcD: electron-transfer-HCD.
In addition to preset options that are fixed for the above modes, gpAnnotate also allows ‘Custom’ definition of fragmentation rules in order to accommodate additional user preferences. Thus, in principle, it can handle a wide variety of scenarios.
A key feature of the program is the use of DrawGlycan-SNFG (version 2, [ref. 1]) for the annotation of MS/MS spectrum. Many of the scoring algorithms incorporated into gpAnnotate are derived from the GlycoProteomics Analysis ToolBox (GlycoPAT, [ref. 2]). Additional updates to GlycoPAT for large scale glycoproteomics studies will be released shortly (manuscript in preparation).
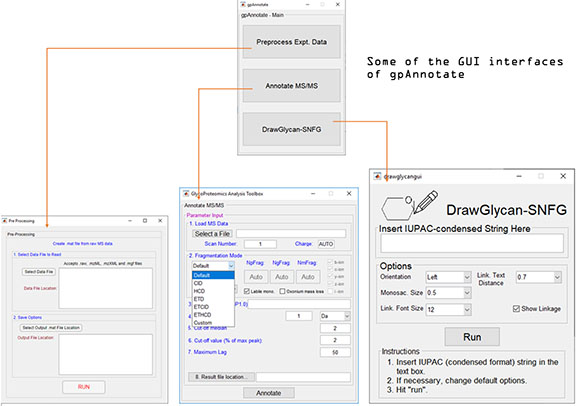
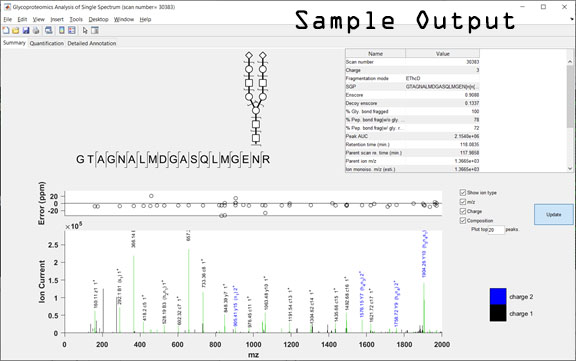
gpAnnotate includes 3 modules. The main GUI (graphical user interface) interface is shown below along with the interfaces for: i. Preprocessing of experimental data; ii. Annotating of MS/MS spectrum; and iii. DrawGlycan-SNFG (version 2). A sample output GUI is also shown.
Instructions for software installation, technical details and usage directions are provided in the program manual. A separate website is also available for DrawGlycan-SNFG.
To test gpAnnotate:
Download the gpAnnotate standalone application and example files using the above link.
i. Select one of the example data files (either “testdata_cryo.mat” or “testdata_mix.mat”);
ii. Copy scan number and glycopeptide SGP sequence from “Example_Candidates_and_Results.xlsx” to the corresponding boxes in the software interface;
iii. Hit “Annotate” to run the program.
References:
1. Cheng, K., Y. Zhou, and S. Neelamegham, DrawGlycan-SNFG: a robust tool to render glycans and glycopeptides with fragmentation information. Glycobiology, 2017. 27(3): p. 200-205.
2. Liu, G., et al., A Comprehensive, Open-source Platform for Mass Spectrometry-based Glycoproteomics Data Analysis. Mol Cell Proteomics, 2017. 16(11): p. 2032-2047.
